Nipype: Difference between revisions
(→Pydra) |
|||
| (10 intermediate revisions by 3 users not shown) | |||
| Line 46: | Line 46: | ||
{{Term|location=frontend.site|cmd=<code class="command">pip install nipype</code>}} | {{Term|location=frontend.site|cmd=<code class="command">pip install nipype</code>}} | ||
== Basic usage == | |||
Let's assume that you have previously installed Nipype's dependencies. | |||
Here, we present an basic example of performing an addition using Nipype's standalone Node: | |||
<syntaxhighlight lang="python"> | |||
from nipype import Node | |||
from nipype.interfaces import Function | |||
def add_two(x): | |||
return x + 2 | |||
add_func = Function(input_names=["x"], output_names=["out"], function=add_two) | |||
add_node = Node(add_func, name="add_node") | |||
add_node.inputs.x = 3 | |||
res = add_node.run() | |||
print(res.outputs.out) | |||
</syntaxhighlight> | |||
== Basic usage == | In the result of the workflow, we can see: | ||
<syntaxhighlight lang="bash"> | |||
231129-15:04:05,941 nipype.workflow INFO: | |||
[Node] Setting-up "add_node" in "/tmp/tmppx3r537_/add_node". | |||
231129-15:04:05,946 nipype.workflow INFO: | |||
[Node] Executing "add_node" <nipype.interfaces.utility.wrappers.Function> | |||
231129-15:04:06,9 nipype.workflow INFO: | |||
[Node] Finished "add_node", elapsed time 0.004518s. | |||
5 | |||
</syntaxhighlight> | |||
== Basic neuroimaging usage == | |||
Let's assume that you have previously installed Nipype's dependencies and you have installed [https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/FslInstallation/Linux fsl] to process the [https://openneuro.org/datasets/ds000003/versions/1.0.0 Rhyme judgment dataset] available on OpenNeuro. | Let's assume that you have previously installed Nipype's dependencies and you have installed [https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/FslInstallation/Linux fsl] (the full installer is [https://git.fmrib.ox.ac.uk/fsl/conda/installer here]) to process the [https://openneuro.org/datasets/ds000003/versions/1.0.0 Rhyme judgment dataset] available on OpenNeuro. | ||
Here, we present an basic example of performing pre-processing using Nipype: | Here, we present an basic example of performing pre-processing using Nipype: | ||
| Line 96: | Line 131: | ||
<syntaxhighlight lang="python"> | <syntaxhighlight lang="python"> | ||
preprocessing.run(plugin=' | preprocessing.run(plugin='OAR', plugin_args=dict(oarsub_args='-q default')) | ||
</syntaxhighlight> | </syntaxhighlight> | ||
| Line 106: | Line 141: | ||
<syntaxhighlight lang="python"> | <syntaxhighlight lang="python"> | ||
mcflirt.plugin_args = {'overwrite': True, 'oarsub_args': '-q production'} | |||
</syntaxhighlight> | </syntaxhighlight> | ||
The basic addition example above can be executed using OAR resources as follows: | |||
<syntaxhighlight lang="python"> | |||
from nipype import Node, Workflow | |||
from nipype.interfaces import Function | |||
from pathlib import Path | |||
def add_two(x): | |||
return x + 2 | |||
add_func = Function(input_names=["x"], output_names=["out"], function=add_two) | |||
add_node = Node(add_func, name="add_node") | |||
add_node.inputs.x = 3 | |||
wf = Workflow(name="basic_wf", base_dir=Path.cwd()) | |||
wf.add_nodes([add_node]) | |||
res = wf.run(plugin='OAR', plugin_args=dict(oarsub_args='-q default')) | |||
print(list(res.nodes())[0].result.outputs) | |||
assert list(res.nodes())[0].result.outputs.out == 5 | |||
</syntaxhighlight> | |||
In the result of the workflow, we can see: | |||
<syntaxhighlight lang="bash"> | |||
240313-17:56:53,738 nipype.workflow INFO: | |||
Workflow basic_wf settings: ['check', 'execution', 'logging', 'monitoring'] | |||
240313-17:56:53,757 nipype.workflow INFO: | |||
Running in parallel. | |||
240313-17:56:54,88 nipype.workflow INFO: | |||
Pending[0] Submitting[1] jobs Slots[inf] | |||
240313-17:56:54,89 nipype.workflow INFO: | |||
Submitting: basic_wf.add_node ID: 0 | |||
240313-17:56:56,914 nipype.workflow INFO: | |||
Finished submitting: basic_wf.add_node ID: 0 | |||
240313-17:57:29,844 nipype.workflow INFO: | |||
[Job 0] Completed (basic_wf.add_node). | |||
out = 5 | |||
</syntaxhighlight> | |||
Well the basic neuroimaging example above can be executed using OAR resources as follows: | |||
<syntaxhighlight lang="python"> | <syntaxhighlight lang="python"> | ||
| Line 128: | Line 207: | ||
# Workflow execution | # Workflow execution | ||
preprocessing.run(plugin='OAR', plugin_args= | preprocessing.run(plugin='OAR', plugin_args=dict(oarsub_args='-q production')) | ||
</syntaxhighlight> | </syntaxhighlight> | ||
Note the use of <code>base_dir</code> here. If no location is specified, Nipype will creates a temporary folder in the /tmp directory, which is not shared across remote resources. That's why we specify an NFS server here as <code>base_dir</code>, to be sure that computations are correctly performed on distant nodes. You should probably check the fsl installation and the input folder also. | {{Warning|text=Note the use of <code>base_dir</code> here. If no location is specified, Nipype will creates a temporary folder in the /tmp directory, which is not shared across remote resources. That's why we specify an NFS server here as <code>base_dir</code>, to be sure that computations are correctly performed on distant nodes. You should probably check the fsl installation and the input folder also.}} | ||
In the result of the workflow, we can see: | In the result of the workflow, we can see: | ||
| Line 155: | Line 234: | ||
Pydra rewrites Nipype engine with mapping and joining as first-class operations. | Pydra rewrites Nipype engine with mapping and joining as first-class operations. | ||
However, Pydra does not have OAR Support yet. | However, the upstream of Pydra does not have OAR Support yet. | ||
Examples and details of our attempt at creating a Pydra's OAR extension can be found [[User:Apetit/Pydra|here]], but this effort is currently stalled. If you would be a user of this, please let us know. | |||
'''Edit (July 2025)''': Since April 2025, the Pydra project has adopted a new architecture. An OAR worker has been implemented accordingly and is available starting from version 1.0a1, see [https://github.com/ychiat35/pydra/tree/feature/oar this branch]. | |||
You can either install it from source, or use the OAR support as an external Pydra plugin, available [https://github.com/ychiat35/pydra_oar here]. | |||
Latest revision as of 16:27, 3 July 2025
| Note | |
|---|---|
This page is actively maintained by the Grid'5000 team. If you encounter problems, please report them (see the Support page). Additionally, as it is a wiki page, you are free to make minor corrections yourself if needed. If you would like to suggest a more fundamental change, please contact the Grid'5000 team. | |
Nipype
What is Nipype?
|
|
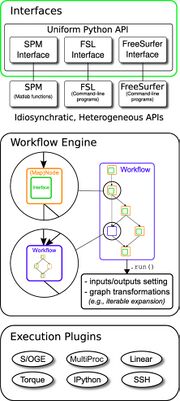
Nipype (Neuroimaging in Python Pipelines and Interfaces) is a flexible, lightweight and extensible neuroimaging data processing framework in Python. It is a community-developed initiative under the umbrella of Nipy. It addresses the heterogeneous collection of specialized applications in neuroimaging: SPM in MATLAB, FSL in shell, and Nipy in Python. A uniform interface is proposed to facilitate interaction between these different packages within a single workflow. The source code, issues and pull requests can be found here. The fundamental parts of Nipype are Interfaces, the Workflow Engine and the Execution Plugins, as you can see in the figure at the left:
Among the execution plugins, you can find |
Installation
pip can be used to install the stable release of Nipype:
It is recommended to install python dependencies within a virtual environment. To do so, execute the following commands before running the pip command:
Basic usage
Let's assume that you have previously installed Nipype's dependencies.
Here, we present an basic example of performing an addition using Nipype's standalone Node:
from nipype import Node
from nipype.interfaces import Function
def add_two(x):
return x + 2
add_func = Function(input_names=["x"], output_names=["out"], function=add_two)
add_node = Node(add_func, name="add_node")
add_node.inputs.x = 3
res = add_node.run()
print(res.outputs.out)
In the result of the workflow, we can see:
231129-15:04:05,941 nipype.workflow INFO:
[Node] Setting-up "add_node" in "/tmp/tmppx3r537_/add_node".
231129-15:04:05,946 nipype.workflow INFO:
[Node] Executing "add_node" <nipype.interfaces.utility.wrappers.Function>
231129-15:04:06,9 nipype.workflow INFO:
[Node] Finished "add_node", elapsed time 0.004518s.
5
Basic neuroimaging usage
Let's assume that you have previously installed Nipype's dependencies and you have installed fsl (the full installer is here) to process the Rhyme judgment dataset available on OpenNeuro.
Here, we present an basic example of performing pre-processing using Nipype:
import nipype.interfaces.fsl as fsl
from nipype import Node, Workflow
preprocessing = Workflow(name='preprocessing')
# using fMRI's linear image registration tool for intra-modal motion correction
mcflirt = Node(fsl.MCFLIRT(), name='mcflirt')
mean = Node(fsl.MeanImage(), name='mean')
# add Nodes to Workflow
preprocessing.connect(mcflirt, 'out_file', mean, 'in_file')
mcflirt.inputs.in_file = '/home/ychi/nipype/ds000003/sub-01/func/sub-01_task-rhymejudgment_bold.nii.gz'
# Workflow execution
preprocessing.run()
In the result of the workflow, we can see:
Running serially- the output folder and the execution time for the mcflirt
- the output folder and the execution time for the mean
Using Nipype Plugins
As shown in the figure above, Nipype's workflow engine supports a plugin architecture for workflow execution. The available plugins, such as SGE, PBS, HTCondor, LSF, Slurm or OAR allow local and distributed execution of workflows and debugging.
All plugins can be executed by calling:
workflow.run(plugin=PLUGIN_NAME, plugin_args=ARGS_DICT)
Using Nipype's OAR plugin
To use the OAR plugin, you need to call:
preprocessing.run(plugin='OAR')
Note: you can also provide traditional oarsub arguments by using the oarsub_args parameter:
preprocessing.run(plugin='OAR', plugin_args=dict(oarsub_args='-q default'))
Other parameters supported by OAR plugin are:
template: custom template file 'hello-world.sh' for batch job submissionmax_jobname_len: maximum length of the job name. Default 15.
Note that plugin_arg of the node level configuration can overwrite the general configuration defined for all the other nodes of the same workflow.
mcflirt.plugin_args = {'overwrite': True, 'oarsub_args': '-q production'}
The basic addition example above can be executed using OAR resources as follows:
from nipype import Node, Workflow
from nipype.interfaces import Function
from pathlib import Path
def add_two(x):
return x + 2
add_func = Function(input_names=["x"], output_names=["out"], function=add_two)
add_node = Node(add_func, name="add_node")
add_node.inputs.x = 3
wf = Workflow(name="basic_wf", base_dir=Path.cwd())
wf.add_nodes([add_node])
res = wf.run(plugin='OAR', plugin_args=dict(oarsub_args='-q default'))
print(list(res.nodes())[0].result.outputs)
assert list(res.nodes())[0].result.outputs.out == 5
In the result of the workflow, we can see:
240313-17:56:53,738 nipype.workflow INFO:
Workflow basic_wf settings: ['check', 'execution', 'logging', 'monitoring']
240313-17:56:53,757 nipype.workflow INFO:
Running in parallel.
240313-17:56:54,88 nipype.workflow INFO:
Pending[0] Submitting[1] jobs Slots[inf]
240313-17:56:54,89 nipype.workflow INFO:
Submitting: basic_wf.add_node ID: 0
240313-17:56:56,914 nipype.workflow INFO:
Finished submitting: basic_wf.add_node ID: 0
240313-17:57:29,844 nipype.workflow INFO:
[Job 0] Completed (basic_wf.add_node).
out = 5
Well the basic neuroimaging example above can be executed using OAR resources as follows:
import nipype.interfaces.fsl as fsl
from nipype import Node, Workflow
preprocessing = Workflow(name='preprocessing', base_dir='/home/ychi/nipype')
# using fMRI's linear image registration tool for intra-modal motion correction
mcflirt = Node(fsl.MCFLIRT(), name='mcflirt')
mean = Node(fsl.MeanImage(), name='mean')
# add Nodes to Workflow
preprocessing.connect(mcflirt, 'out_file', mean, 'in_file')
mcflirt.inputs.in_file = '/home/ychi/nipype/ds000003/sub-01/func/sub-01_task-rhymejudgment_bold.nii.gz'
# Workflow execution
preprocessing.run(plugin='OAR', plugin_args=dict(oarsub_args='-q production'))
In the result of the workflow, we can see:
Running in parallel- the output folder and the execution time for the mcflirt
- the output folder and the execution time for the mean
Using Nipype's Slurm plugin
For example, to execute the basic preprocessing example above with Slurm resources, you need to call:
preprocessing.run(plugin='SLURM')
Other parameters supported by Slurm plugin are:
sbatch_args: command line args to be passed to sbatchtemplate: custom template file 'hello-world.sh' for batch job submission
Pydra
Pydra is a part of the second generation of the Nipype ecosystem, which is meant to provide additional flexibility and reproducibility.
Pydra rewrites Nipype engine with mapping and joining as first-class operations.
However, the upstream of Pydra does not have OAR Support yet.
Examples and details of our attempt at creating a Pydra's OAR extension can be found here, but this effort is currently stalled. If you would be a user of this, please let us know.
Edit (July 2025): Since April 2025, the Pydra project has adopted a new architecture. An OAR worker has been implemented accordingly and is available starting from version 1.0a1, see this branch. You can either install it from source, or use the OAR support as an external Pydra plugin, available here.