Nipype
| Note | |
|---|---|
This page is actively maintained by the Grid'5000 team. If you encounter problems, please report them (see the Support page). Additionally, as it is a wiki page, you are free to make minor corrections yourself if needed. If you would like to suggest a more fundamental change, please contact the Grid'5000 team. | |
Nipype
What is Nipype?
Nipype (Neuroimaging in Python Pipelines and Interfaces) is a flexible, lightweight and extensible neuroimaging data processing framework in Python. It is a community-developed initiative under the umbrella of Nipy. It addresses the heterogeneous collection of specialized applications in neuroimaging: SPM in MATLAB, FSL in shell, and Nipy in Python. A uniform interface is proposed to facilitate interaction between these different packages within a single workflow.
The source code, issues and pull requests can be found here.
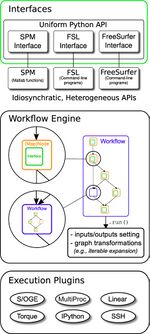
The fundamental parts of Nipype are Interfaces, the Workflow Engine and the Execution Plugins, as you can see in the figure at the left:
- Interface: wraps a program or function
- (Map)Node: wraps an Interface for use in a Workflow
- Workflow: a graph or a forest of graphs whose edges represent data flow
- Plugin: a component which describes how a Workflow should be executed
Among the execution plugs, you can find anOAR plugin here.
Installation
Basic usage
Pydra
Pydra is a part of the second generation of the Nipype ecosystem, which is meant to provide additional flexibility and reproducibility. Pydra rewrites Nipype engine with mapping and joining as first-class operations. However, Pydra does not have OAR Support. Examples and details of Pydra's OAR extension can be found here.